7.2 Furin site
Th spike glycoprotein contains many features (section 6.1). We’ll just take a look at the results for the novel furin recognition site at residue 682. The furin recognition sequence is RRAR
The alignment at this location appears different between our automated TCoffee version and the Walls version.

Figure 7.1: Alignment details around the furin site for the automated TCoffee alignment. Note the structure information at the top line and the consensus sequence at the bottom line.

Figure 7.2: Alignment details around the furin site for Walls (2020) alignment. Note the consensus sequence at the bottom line.
Figure 7.1 depicts the result of our automated alignment and figure 7.2 that of the Walls paper. It is indeed unfortunate, but not unexpected, that this region is not visible (hence absent) from the PDB sequences since the sequences are likely cleaved allowing too much flexibility to the cut ends.
A PyMOL (Schrödinger, LLC (2020)) illustration of this region is shown in figure 7.3. The script used to create the image can be found in appendix B. The inset is simply a zoomed out version of the same. The last visibe residues on each strand are labeled. Residues 677 to 689 have not been resovled.
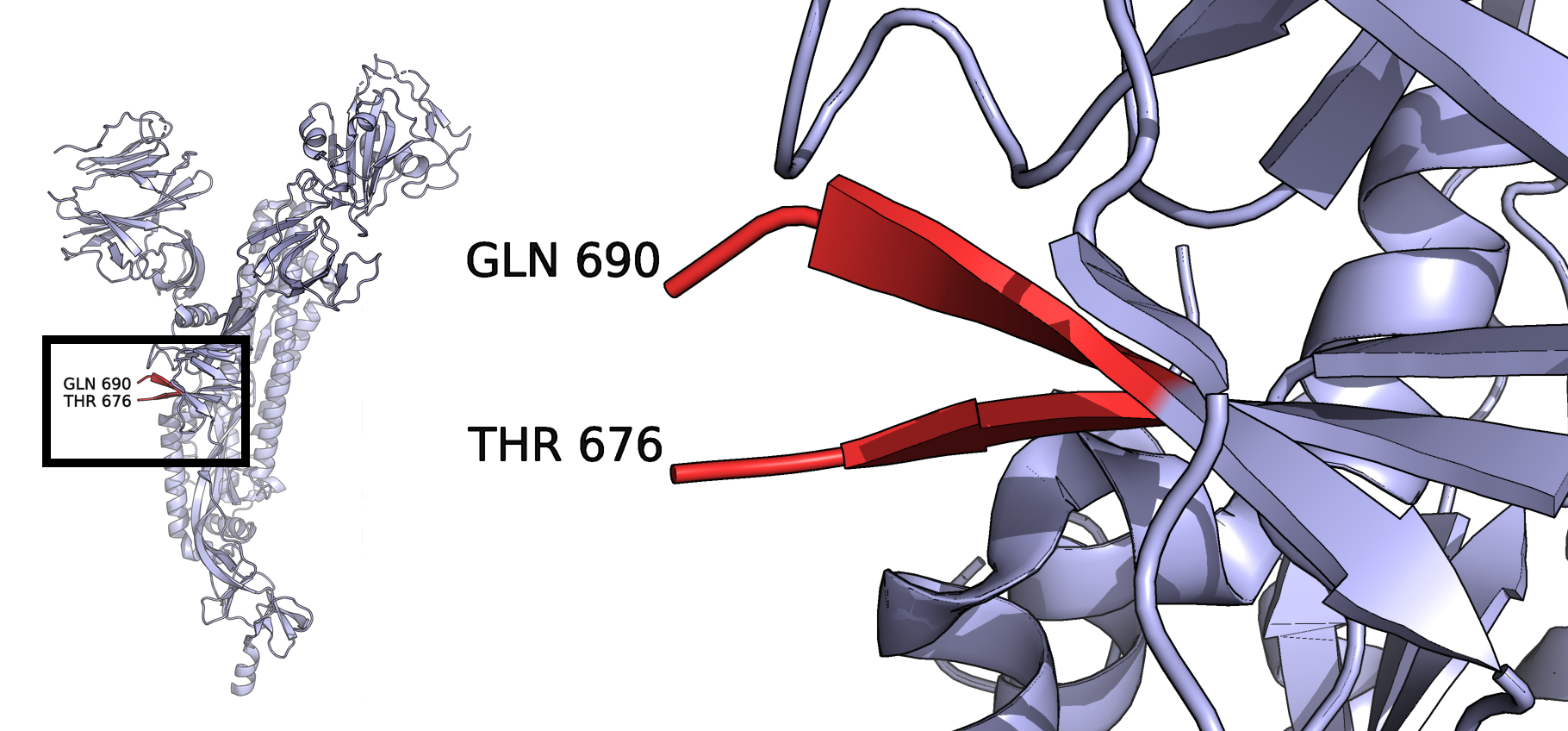
Figure 7.3: PDB ID 6VYB, one chain of the trimeric spike protein showing the missing amino acids around the novel furin cleavage site.
In figure figure 7.2 the four amino acids that appear to be extra above a column of dots are PRRA while in figure 7.1 they appear as NSPR.
In addition, a TCoffee Expresso run on the web site a few days ago gave a slight different result in this area as well, the “floating” four amino acids were SPRR.
SARS-CoV-2 641 NVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIA 694
SARSr-CoV_RaTG1 641 NVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTN----SRSVASQSIIA 690
SARS-CoV_Urbani 627 NVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSL----LRSTSQKSIVA 676
SARS-CoV_CUHK-W 627 NVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSL----LRSTSQKSIVA 676
SARS-CoV_GZ02 627 NVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSL----LRSTSQKSIVA 676
SARS-CoV_A031 627 NVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSS----LRSTSQKSIVA 676
SARS-CoV_A022 627 NVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSS----LRSTSQKSIVA 676
WIV16 627 NVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSS----LRSTSQKSIVA 676
WIV1 628 NVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSS----LRSTSQKSIVA 677
SARSr-CoV_ZXC21 617 SVFQTQAGCLIGAEHVNASYECDIPIGAGICASYHTASI----LRSTGQKAIVA 666
SARSr-CoV_ZC45 618 NVFQTQAGCLIGAEHVNASYECDIPIGAGICASYHTASI----LRSTSQKAIVA 667
SARSr-CoV_Rp3 613 NVFQTQAGCLIGAEHVNASYECDIPIGAGICASYHTAST----LRSVGQKSIVA 662
SARSr-CoV_Rs672 613 NVFQTQAGCLIGAEHVNASYECDIPIGAGICASYHTAST----LRSVGQKSIVA 662
cons 649 .****:**********: ****************:* : **...::*:* 702 The 3D structure does not help us resolve these conflicts, but it is rather easy to see that moving 2 columns of amino acids from the automated TCoffee alignment just made or one column of the web Expresso version to the left would reproduce the Walls paper version. This could be accomplished with a manual editor that allow easy editing of alignments such as Jalview.
Overall TCoffee Expresso run on the web (not shown) gave a score of “Good” to most of the sequences over their length providing each of these sequences with a score between 97 and 99 with an average score of 98 out of 100.
References
Schrödinger, LLC. 2020. “The PyMOL Molecular Graphics System, Version 2.0.” https://pymol.org/.