6.1 Spike protein features
Before attempting an alignment of the SARS_CoV-2 spike protein to other similar structure let’s look at a summary of the the spike protein sequence features. Figure 1 of Lokman et al. (2020) provides a graphical comparison between SARS_CoV and SARS-CoV-2 spike sequence features.
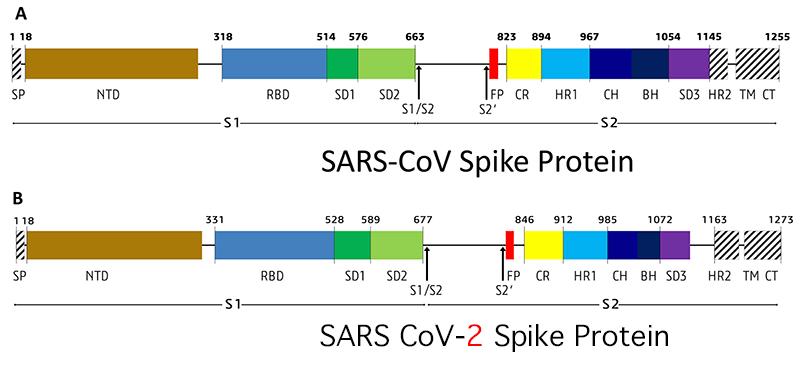
Figure 6.1: SARS-CoV and CoV-2 Spike protein.
Figure 6.1 legend: SP= signal peptide, NTD= N-terminal domain, RBD= receptor-binding domain, SD1= subdomain 1, SD2= subdomain 2, S1/S2= S1/S2 protease cleavage site, S2′= S2′ protease cleavage site, FP= fusion peptide, HR1= heptad repeat 1, CH= central helix, CH= central helix, BH= β-hairpin, HR2= heptad repeat 2, TM= transmembrane domain, CT= cytoplasmic tail. Arrows denote protease cleavage sites.
SARS-CoV-2 has emerged with remarkable properties that include a novel, unique furin cleavage site (PRRAR↓SV) at S1/S2 boundary in the S spike glycoprotein.
The role of the sequence features of the spike protein is elegantly summarized by Lokman et al. (2020):
“Viral entry to the host cell is initiated by the receptor-binding domain (RBD) of S1 head. Upon receptor-binding, proteolytic cleavage occurs at S1/S2 cleavage site and two heptad repeats (HR) of S2 stalk form a six-helix bundle structure triggering the release of the fusion peptide. As it comes into close proximity to the transmembrane anchor (TM), the TM domain facilitates membrane destabilization required for fusion between virus-host membranes.”
References
Lokman, S. M., M. Rasheduzzaman, A. Salauddin, R. Barua, A. Y. Tanzina, M. H. Rumi, M. I. Hossain, A. M. A. M. Z. Siddiki, A. Mannan, and M. M. Hasan. 2020. “Exploring the genomic and proteomic variations of SARS-CoV-2 spike glycoprotein: A computational biology approach.” Infect. Genet. Evol. 84 (June): 104389. https://doi.org/https://doi.org/10.1016/j.meegid.2020.104389.